Introduction
MoonDB contains Extreme Multifunctional (EMF) candidates from the MoonGO pipeline (Chapple et al. (2015a)), which combines network topological information and protein annotations, as well as a manually curated set of human moonlighting proteins collected from the literature.
This database focuses on the functional annotation of these proteins and combines with annotations from UniProt.
MoonDB currently contains mostly data for Human but also for Mouse, Fly, Worm and Yeast.
Tutorial
1. Search a protein entry by UniprotKB ID, AC (accession number) or Gene Name using the search box in the home page.

2. Alternatively, browse all MoonDB proteins. These can be filtered by 'Species', 'Reference Set' or other combinations of columns (case-insensitive search). Explore a protein entry by clicking under the "MoonDB ID" column.
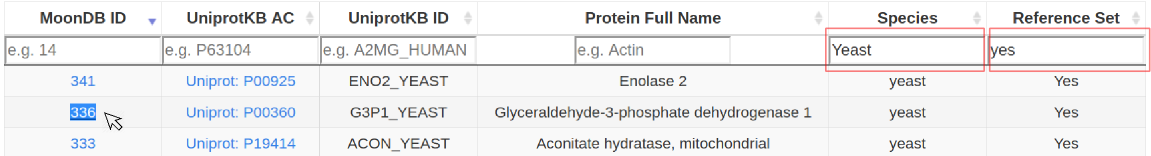
3. Protein entries: different sets of functional annotations, protein interactions, and other relevant information can be expanded and filtered.
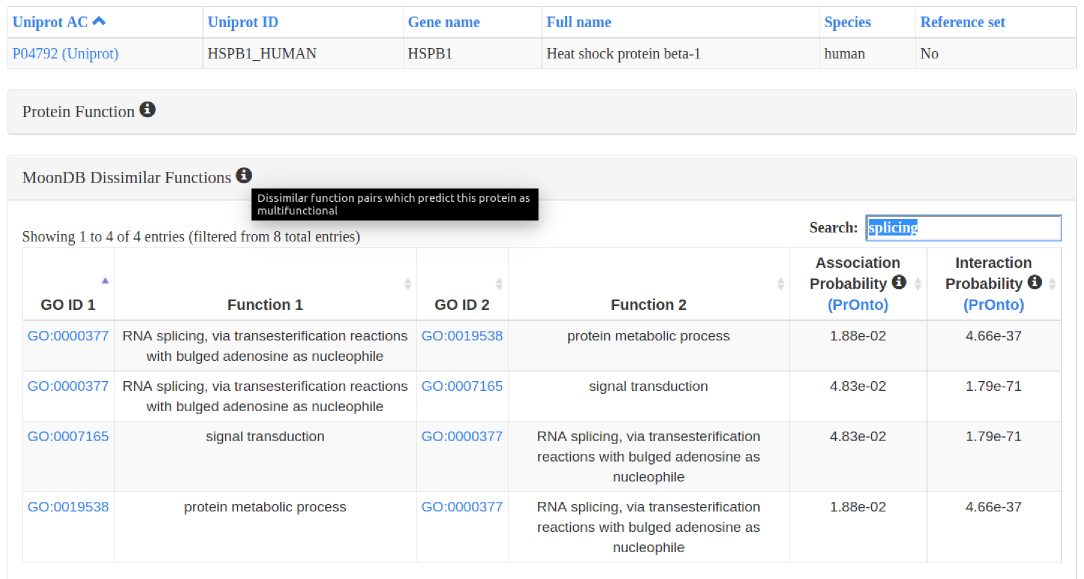
4. Downloads: the lists of MoonDB proteins and their extreme multifunctional functions, as well as the protein interactomes, can be downloaded as tsv files.
Synopsis of the MoonGO pipeline
- Overlapping clusters (network modules) are extracted from a protein-protein interaction network using the OCG algorithm (Becker at al., 2012).
- Clusters are annotated according to the GO annotations (Biological Processes) of their constituent proteins (using a majority rule).
- Extreme Multifunctional protein candidates are identified at the intersection of clusters involved in unrelated biological processes according to PrOnto GO term association probabilities (Chapple et al., 2015b).

MoonGO: the EMF identification pipeline.
PrOnto probabilities
PrOnto determines pairs of GO terms that are functionally dissimilar to each other. The original PrOnto publication can be found here.
The PrOnto probabilities used in MoonDB v2.0 are using the same algorithm, but recalculated for the updated protein interactome and GO term annotations.
Let X be the variable “number of proteins annotated to both terms” which follows a hypergeometric law. The probabilites are calculated with this equation:
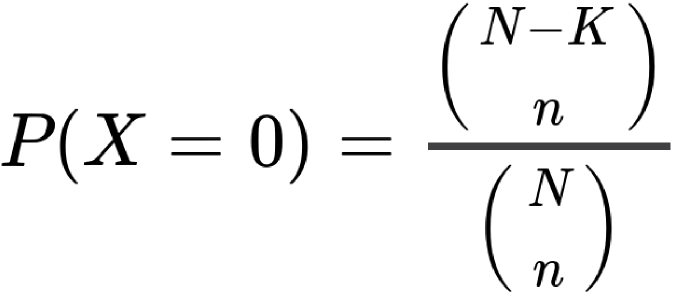

Where, for Association Probability:
- N is the number of proteins with at least one direct annotation to a term of the source ontology of GO1 and one direct annotation to a term of the source ontology of GO2;
- K is the number of proteins directly annotated to GO1;
- n is the number of proteins annotated to GO2;
- k is the number of proteins annotated to both terms.
For the Interaction Probability:
- N the total number of interactions in the query species' network between proteins annotated to at least one term of the source ontology of GO1 and proteins annotated to at least one term of the source ontology of GO2;
- K is the number of interactions involving proteins annotated to GO1;
- n the number of interactions involving proteins annotated to GO2;
- k the the number of interactions between a protein annotated to GO1 and one annotated to GO2.
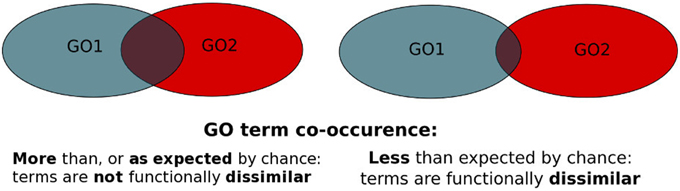
MoonDB datasets
The MoonGO pipeline was used to detect EMF candidates in human (Homo sapiens), mouse (Mus musculus), fruit fly (Drosophila melanogaster), worm (Caenorhabditis elegans) and yeast (Saccharomyces cerevisiae).
As the MoonGO pipeline requires both a high quality protein interactome and extensive GO term annotations, the prediction power for mouse, fly and worm (with either an incomplete interactome or incomplete GO term annotations) is lower than for human and yeast. This means less candidates will be found for species lacking in interactome/annotation quality.
